Retrieve Data from Entry ID
A series of NMR-STAR data formats, NMR-STAR v3.2, BMRB/XML and BMRB/RDF, is available through the next form. Please enter Entry ID and select download links to each files.
This site provides APIs for retrieving BMRB/XML and BMRB/RDF data using Entry ID embedded URI. Please find usage of REST APIs and connect your client tools.
Download using HTTP protocol
NMR-STAR v3 (BMRB):
https://bmrb.pdbj.org/ftp/pub/bmrb/entry_lists/nmr-star3.1/NMR-STAR v3 (BMRB+PDB):
https://bmrb.pdbj.org/ftp/pub/bmrb/nmr_pdb_integrated_data/coordinates_restraints_chemshifts/bmrb_plus_pdb/NMR-STAR v3 (Metabolomics; experimental):
https://bmrb.pdbj.org/ftp/pub/bmrb/metabolomics/NMR_STAR_experimental_entries/NMR-STAR v3 (Metabolomics; theoretical):
https://bmrb.pdbj.org/ftp/pub/bmrb/metabolomics/NMR_STAR_theoretical_entries/BMRB/XML (complete):
- Master (BMRB+Metabolomics)
- https://bmrbpub.pdbj.org/archive/xml/
- BMRB
- https://bmrb.pdbj.org/ftp/pub/bmrb/entry_lists/xml/
- Metabolomics
- https://bmrb.pdbj.org/ftp/pub/bmrb/metabolomics/entry_lists/xml/
BMRB/XML (noatom):
- Master (BMRB+Metabolomics)
- https://bmrbpub.pdbj.org/archive/xml-noatom/
- BMRB
- https://bmrb.pdbj.org/ftp/pub/bmrb/entry_lists/xml-noatom/
- Metabolomics
- https://bmrb.pdbj.org/ftp/pub/bmrb/metabolomics/entry_lists/xml-noatom/
BMRB/RDF (RDF/XML):
- Master (BMRB+Metabolomics)
- https://bmrbpub.pdbj.org/archive/rdf/
- BMRB
- https://bmrb.pdbj.org/ftp/pub/bmrb/entry_lists/rdf/
- Metabolomics
- https://bmrb.pdbj.org/ftp/pub/bmrb/metabolomics/entry_lists/rdf/
BMRB/RDF (N-Triples):
- Master (BMRB+Metabolomics)
- https://bmrbpub.pdbj.org/archive/rdf-nt/
- BMRB
- https://bmrb.pdbj.org/ftp/pub/bmrb/entry_lists/rdf-nt/
- Metabolomics
- https://bmrb.pdbj.org/ftp/pub/bmrb/metabolomics/entry_lists/rdf-nt/
BMRB/JSON (noatom):
Bulk Download using rsync protocol
The following rsync commands are useful for mirroring and updating of BMRB/XML and BMRB/RDF archives, respectively.
BMRB/XML (complete):
% rsync -av --delete rsync://bmrbpub.pdbj.org/bmrb-xml .
BMRB/XML (noatom):
% rsync -av --delete rsync://bmrbpub.pdbj.org/bmrb-xml-noatom .
BMRB/RDF: (RDF/XML)
% rsync -av --delete rsync://bmrbpub.pdbj.org/bmrb-rdf .
BMRB/RDF (N-Triples):
% rsync -av --delete rsync://bmrbpub.pdbj.org/bmrb-rdf-nt .
BMRB/JSON (noatom):
% rsync -av --delete rsync://bmrbpub.pdbj.org/bmrb-json-noatom .
Automated Translation & Remediation Tools
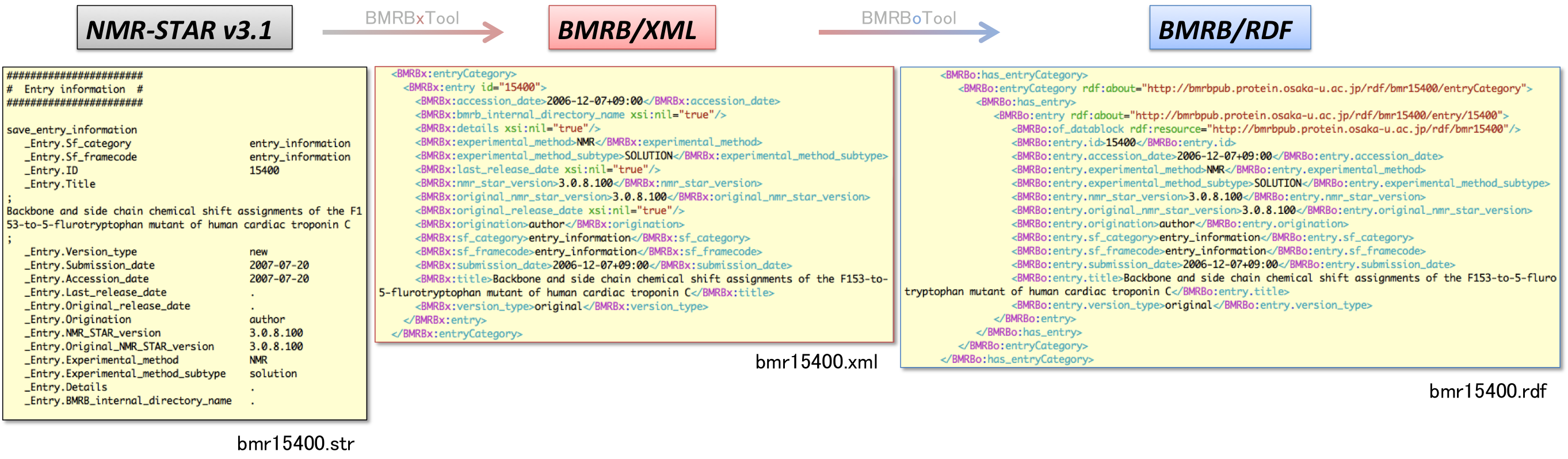
We have developed a suite of conversion tools, BMRBxTool and BMRBoTool. The BMRBxTool generates qualified XML files by programmed remediation, and reports validation errors against the XML schema. Then, the BMRBoTool converts NMR-STAR data (BMRB/XML) into RDF format (BMRB/RDF). By using scripts bundled with the tools, the XML and RDF documents can be updated for both new entries and revised ones.
Translation example of "Entry" category for BMRB entry 15400
nmrML converter for BMRB metabolomics entries
We have developed BMSxNmrML, an nmrML converter for BMRB metabolomics entries based on BMRB/XML. The nmrML is an open mark-up language for NMR data (FID). It utilizes owl-indexer, an OWL literal indexing tool, for identifying chemical compounds from ChEBI OWL.
Database Replication Tool
We have developed a database replication tool based on XML Schema, xsd2pgschema, whose executable jar file (xsd2pgschema.jar) has been bundled with the BMRBxTool. The xsd2pgschema is Java application suite, which converts XML Schema 1.1 to PostgreSQL DDL and supports migrating XML data into PostgreSQL DB based on the XML Schema without defects on information content. Source of program is provided under Apache License V2.0.
OWL Literal Indexing Tool
We have developed a full-text index generator of OWL literal, owl-indexer, The owl-indexer generate full-text index of OWL literal via either Apache Lucene or Sphinx Search. It is based on OWL API. Source of program is provided under Apache License V2.0.



